Computational Analysis and Prediction of Phosphopeptide Binding Sites
Brian A. Joughin, Michael B. Yaffe & Bruce Tidor
A machine-learning technique has been developed for the purpose of analyzing the properties of phosphopeptide binding sites on the surface of proteins, and was applied predictively to the surface of a pair of phosphopeptide binding domains for which the ligand binding sites were not crystallographically determined [5]. This technique focuses on the local chemical and physical properties of extremely small portions of the protein surface. Predictions made by this technique have been validated by experiment.
Motivation
Protein phosphorylation leads to the regulation of signaling, often by the generation of a binding site for a phosphopeptide binding domain. Though all phosphopeptide binding domains are capable of binding to phosphorylated peptides, there is relatively little structural similarity in the binding sites. An understanding of the commonalities among the natural phosphopeptide binding domains should be useful in continuing efforts to design new phosphopeptide binding domains with novel specificity. Moreover, an understanding of what constitutes a phosphopeptide binding site lends predictive ability in cases where a protein of known structure is known to function as a phosphopeptide binding domain at an unknown surface location.
Methods
A set of nine crystal structures of phosphopeptide binding domains in complex with phosphopeptides were surfaced with a triangulated mesh. At each mesh vertex, a set of physical and chemical properties including amino acid identity, local surface curvature, and solvated electrostatic potential were calculated. The enrichment of these properties in sites which are bound to phosphorylated amino acid side chains with respect to the entire protein surface was determined. In order to determine the predictive capacity of this propensity information, a jack-knifing validation procedure was used in which each crystal structure was removed from the training data, and propensities were recalculated. The learned propensities were then painted onto the surface of the removed protein, using the assumption that propensities based on the three characteristics studied combine independently. The propensities learned from the entire training set were applied in the same way to the surface of BRCA1, which was recently identified as a phosphopeptide binding domain, but for which binding location data was unknown [7,11], as well as to the surface of the checkpoint kinase Chk1, for which only non-crystallographic evidence of the site of phosphopeptide binding existed.
Results
Visual inspection of the results of jack-knife validation indicates that the current surface-element model of phosphopeptide binding is predictive, with little tendency to false negative predictions, but somewhat higher tendency to give false positives. Since this allows the generation of experimentally testable hypotheses, it is significantly more useful than the converse.
Such hypotheses were generated in the case of the BRCT domain of the protein BRCA1 and the kinase Chk1. Mutations to the protein BRCA1, including one which abrogates binding to phosphopeptides [7] are commonly associated with breast and ovarian cancer in women. When the model developed here was applied to the surface of the rat BRCA1 structure [5], two putatitve phosphopeptide binding sites were found. One of these was crystallographically determined to be a correct prediction of the site of phosphopeptide binding [2,9,10]. Likewise, two potential binding sites were found on the surface of Chk1, and experimental evidence exists to indicate that one of them may be the correct site [3].
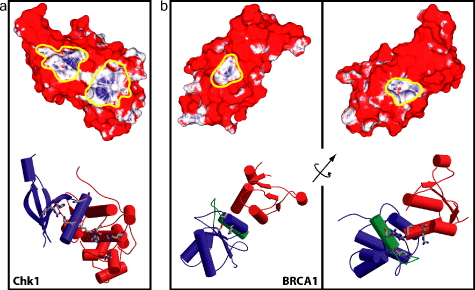
Figure 1: Computed phosphoresidue contact
propensity on the surfaces of Chk1 and BRCA1. In upper panels, blue coloration
indicates high computed propensity for phosphoresidue contact, while red
coloration indicates a low propensity. Yellow outlines indicate predicted
phosphopeptide binding sites. Lower panels show the backbone structures
of the proteins, and were generated using the programs MOLSCRIPT [6] and
RASTER3D [8]. (a) Chk1 (PDB Code 1IA8 [1]). (b) BRCA1
(PDB Code 1L0B [2]).
Future
Model improvements are being considered in which non-phosphopeptide ligand binding propensities are calculated and used to distinguish phosphopeptide-binding sites from sites which are more generically "sticky", as this is not selected against explicitly in the current model and may contribute to the rate of false positives. This would perhaps allow our method to be used in a prospective fashion to mine the Protein Data Bank for novel phosphopeptide binding domains.
Research Support
This work was partially supported by the National Institutes of Health.
References
[1] P. Chen, C. Luo, Y. L. Deng, K. Ryan, J. Register, S. Margosiak, A. Tempczyk-Russel, B. Nguyen, P. Myers, K. Lundgren, er al. The 1.7 Å crystal structure of human cell checkpoint kinase Chk1: Implications for Chk1 regulation. Cell, 100:681-692, 2000.
[2] J. A. Clapperton, I. A. Manke, D. M. Lowery, T. Ho, L. F. Haire, M. B. Yaffe, and S. J. Smerdon. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat. Struct. Mol. Biol., 11:512-518, 204
[3] S. Y. Jeong, A. Kumagai, J. Lee, and W. G. Dunphy. Phosphorylated claspin interacts with a phosphate-binding site in the kinase domain of Chk1 during ATR-mediated activation. J. Biol. Chem., 278:46782-46788, 2003.
[4] W. S. Joo, P. D. Jeffrey, S. B. Cantor, M. S. Finnin, D. M. Livingston, and N. P. Pavletich. Structure of the 53BP1 region bound to p53 and its comparison to the Brca1 BRCT structure. Genes and Development, 16(5):583--593, 2002.
[5] Brian A. Joughin, Bruce Tidor, and Michael B. Yaffe. A computational method for the analysis and prediction of protein:phosphopeptide-binding sites. Protein Science, 14(1):131-139, 2005
[6] P. J. Kraulis. Molscript -- A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24:946-950, 1991.
[7] Isaac A. Manke, Drew M. Lowery, Anhco Nguyen, and Michael B. Yaffe. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science, 302:636--639, 2003.
[8] E. A. Merritt and D. J. Bacon. Raster3d: Photorealistic molecular graphics. Methods Enxymol., 277:505-524, 1997.
[9] E. N. Shiozaki, L. Gu, N. Yan, and Y. Shi. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: Implications for signalling. Mol. Cell., 14:405-412, 2004.
[10] R. S. Williams, M. S. Lee, D. D. Hau, and J. N. Glover. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat. Struct. Mol. Biol., 11:519-525, 2004.
[11] Xiaochun Yu, Claudia Christiano Silva Chini, Miao He, Georges Mer, and Junjie Chen. The BRCT domain is a phospho-protein binding domain. Science, 302:639--642, 2003.
The Stata Center, Building 32 - 32 Vassar Street - Cambridge, MA 02139 - USA tel:+1-617-253-0073 - publications@csail.mit.edu (Note: On July 1, 2003, the AI Lab and LCS merged to form CSAIL.) |
