The Creation of Cytosine Phosphorybosyltransferase
Surasak Chunsrivirot, Michael D. Altman, James R. Williamson & Bruce Tidor
What
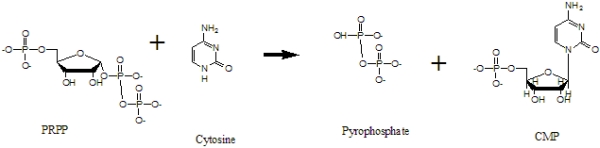
Pyrimidine/purine phosphoribosyl transferases are enzymes that catalyze the formation of a pyrimidine or purine nucleotide by salvage of the preformed base. Nucleotide and pyrophosphate are produced from the base and phosphoriboyslpyrophosphate (PRPP). The goal of this project is to redesign the active site of a purine or pyrimidine phosphoribosyltransferase to create "Cytosine Phosphoribosyltransferase" that can catalyze the formation of cytidine 5'-monophosphate (CMP) from cytosine and PRPP. No natural enzyme that catalyzes this reaction has been found to date, but we plan to engineer an enzyme that performs this important chemistry. The formation of CMP from PRPP and cytosine is shown in Figure 1.
Figure 1. The formation of CMP from PRPP and cytosine
The usefulness of such an enzyme is for the incorporation of labeled cytosine (C) into RNA. Since the enzyme that catalyzes the formation of CMP directly does not exist, we need to redesign an existing enzyme to produce CMP, and this CMP will ultimately be incorporated into RNA.
How
Structure Selection
To create Cytosine Phosphoribosyl Transferase, we need promising crystal structures of Pyrimidine/Purine Phosphoribosyltransferases to use as base structures in our design approach. We used the following criteria in structure selection: a structure should contain the substrates (base ring, PRPP) and cofactors (metal ions); it should contain side-chain interactions (H-bonds) with the base; the base should have functional groups that are similar to cytosine; the active site should be enclosed by a loop. After exploring structures deposited in the Protein Data Bank, we selected two structures: 1TC2 (hypoxanthine phosphoribosyltransferase) and 1L1R (adenine phosphoribosyltransferase).
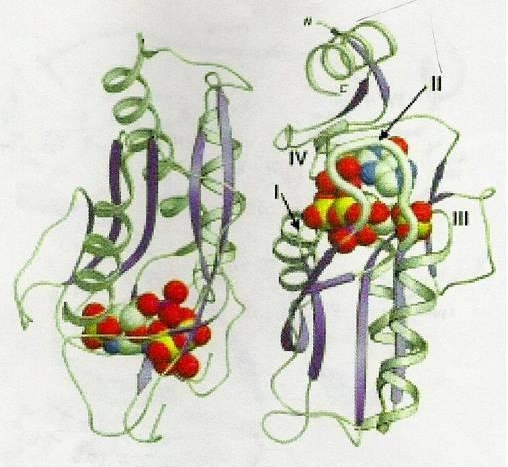
1TC2 is a hypoxanthine phosphoribosyltransferase that catalyzes the formation of inosine 5'-monophosphate (IMP) from hypoxanthine and phosphoribosylpyrophosphate (PRPP); it is an asymmetric dimer from the protozoan parasite Trypanosoma cruzi. This structure contains PRPP, an analogue of the substrate (7-hydroxy[4,3-d]pyrazolopyrimidine) (HPP), Mg & Mn ions. It has one open and one closed subunit; the active site is located between the core and hood domains of the enzyme. This enzyme has a flexible loop closeing the active site upon binding with substrates. The structure of 1TC2 is shown in Figure 2.
Figure 2. The structure of 1TC2, crystallized with a dimer in the asymmetric unit. The monomer on the right has the closed flexible loop (II) that is highlighted as a thicker rendition. This enzyme has the active site located between the core and hood domains; the active site is surrounded by four loops (I-IV). In this figure, HPP,PRPP and hydrated metal ions are shown in standard atom cloloring of van der Waals representation [Figure reproduced from reference [1]]
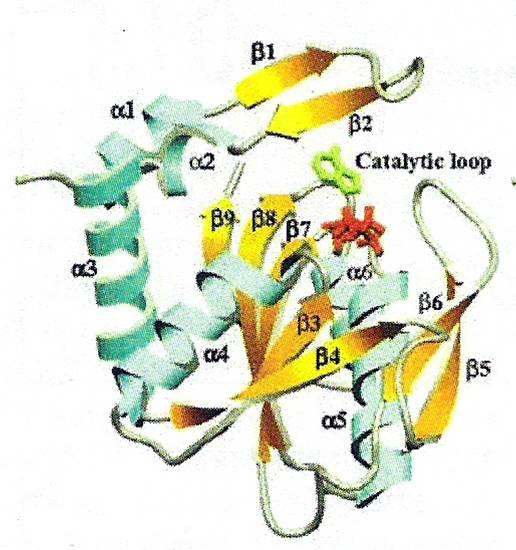
1L1R is an Adenine Phosphoribosyltransferase that catalyzes the formation of AMP from Adenine and PRPP. It is a symmetric homodimer. The hood is composed of alpha 1, 2 and beta 1, 2; the core consists of the remaining alpha helices and beta strands.The catalytic residue is on the loop over the active site pocket, and the loop will be closed when the substrate binds to the active site. This enzyme was co-crystalized with 9-deazaadenine (9DA), Mg ion, PRPP, and its sturcture is shown in Figure 3.
Figure 3. The structure of 1L1R. The monomer of 1L1R has six alpha-helices and nine beta-strands; the enzyme's catalytic loop (beta5 and beta6) is closed onto the active site. 9dA is shown in green, and PRPP is shown in red. [Figure reproduced from reference [2]]
Design Approach
Since enzymes, naturally, are good at binding a transition state of a substrate so that they can catalyze the formation of the product effectively, we proposed the resonance structures of cytosine that are likely to be the transition states by proposing two possible mechanisms (resulting in two resonance forms of cytosine). We built these resonance forms into the active sites of 1TC2 and 1L1R in the same position and orientation as those of existing base, and we used these structures as inputs of the software. Moreover, we selected the designed positions and possible amino acids at a specific position based on the knowledge from literature and from molecular modeling.
Since there are more than thousands of possible sequences resulting from the output of the software, we used the following criteria in selecting promising sequences. The output structures should have a new catalytic residue that is close enough to abstract a proton from the cytosine resonance, a residue that can make hydrogen bonds to -NH2 of cytosine, an aromatic residue that is in the same plane and close to the base ring of cytosine and residues that are able to fill in spaces that the six-membered ring part of the purine base used to reside.
Progress
We have completed two designs on 1TC2 and 1L1R (each with one resonance form of cytosine) and selected promising structures/sequences. Now, we are trying to make a library of sequences that has as high percent coverage as possible.
Research Support
This research was supported by MIT's Undergraduate Reseach Opportunities Program and the John Reed Fund.
References
[1] P. J Focia, S. P. Craig III and A. E. Eakin: Approaching the Transition State in the Crystal Structure of a Phosphoribosyltransferase. In Biochemistry, 37, pp. 17120,1998
[2]W. Shi, Anne Sarver, Ching Wang, Kelly Tanaka, Kelly and Steven Almo. Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia Lamblia Reveal a Mechanism of Ribosyl Migration. In J.Biol.Chem., 277 pp. 39981, 2002
[3]S. P. Craig III and A. E. Eakin. Purine Phosphoribosyltransferases. In The Journal of Biological Chemistry 275, pp. 20231-20234, 2000
[4]Kadziola, Anders., Neuhard, Jan & Larsen, Sine. Structure of product-bound Bacillus caldolyticus uracil phosphoribosyltransferase confirms ordered sequential substrate binding. In Biological Crystallography, D58, pp. 936-945, 2002.
The Stata Center, Building 32 - 32 Vassar Street - Cambridge, MA 02139 - USA tel:+1-617-253-0073 - publications@csail.mit.edu (Note: On July 1, 2003, the AI Lab and LCS merged to form CSAIL.) |