Efficient Population Registration of 3D Data
L. Zöllei, W. Wells & E. Learned-Miller
Introduction
In this project we examine the problem of aligning sets of image volumes where the number of inputs is greater than 20. We call such a task as population alignment. More specifically, we aim to identify a set of homologies that would put the coordinate systems of a large number of medical input volumes into correspondence. One example data set whose elements are to be aligned is displayed on Figure 1, where a central slice from all the input volumes of a baby brain magnetic resonance (MRI) data set is displayed. We believe that defining a robust solution for such an inter-subject registration technology will allow us to build better structural atlases, and to further analyze inter-subject differences. We demonstrate a new framework for aligning populations of medical image volumes for the purpose of digital anatomical atlas construction.
Although our examples in the following are all from the medical domain, we emphasize that the algorithm formulation is very general and it does not contain any specific assumptions about the nature of the input data.
Background
Several approaches exist that propose the alignment of multiple data sets into the same coordinate frame. Besides the details of the registration algorithm applied, there is a significant difference in how they all interpret the coordinate frame or the template with which all elements should be aligned. For some specific applications, this template already exists, for example, as a result of some manual segmentation. The data sets then can be just aligned with the reference frame individually. This approach is advantageous only if the data sets are presented case by case, successively in time.
For other applications the digital template is not available, so that too has to be generated along with the aligning transformations. One group of algorithms selects a standard coordinate frame (for example, based upon certain anatomical structures) and requires the algorithm to position all the inputs of interest into that. The mean of the so-aligned images is then computed. Other approaches select one of the current data points to be the common reference frame. After all the other images are aligned to this, a mean image is computed. Major disadvantages of these methods are that the images need to be pre-processed and the matching landmarks need to be reliably located which is a time-consuming and potentially error-prone procedure. Significant bias can also be introduced by claiming that one data point can represent the standard reference.
There is also growing interest in generating mean models as a by-product of a larger-scale registration process. That formulation eliminates the risk of introducing bias into the registration by simultaneously evolving the data sets towards a common reference. Our approach is one of those.
Our Method
We use a technique called congealing as a basis of our alignment. In that framework, a model of the central tendency of the inputs is derived through an entropy minimization procedure. More specifically, it is the total sum of voxel-wise entropies of the joint image that is to be optimized. The main intuition behind such a formulation is that when in proper alignment, intensity values at corresponding coordinate locations from all the inputs form a low entropy distribution.
Our contribution to the congealing framework lies in its adaptation to a population of grayscale-valued 3D data volumes and its implementation via a stochastic gradient-based optimization procedure. In order to avoid getting trapped in local optima and to improve computation speed, we implemented a multi-resolution framework. It starts the processing of the data sets at a down-sampled and smoothed level and then refines the results during the higher resolution iterations. The number of hierarchy levels is mostly dependent on the quality of the input images and also its size. For our experiments, it was sufficient to use only two levels of hierarchy.
Experiments
We describe two sets of experiments that demonstrate the key properties and the performance characteristics of our algorithm. The total running time was between 30 minutes to 1.5 hour (depending on number of inputs and number of hierarchy levels constructed).
Synthetic
The first experiment is run on a synthetic population. One particular medical MRI volume was selected and a database of transformed volumes was created by applying affine transformations to it. The magnitude of these transformations varied between 0-40 degrees for rotation, 0-40 mm for displacement and between [.5, 1.5] factors for scaling. At the onset of the algorithm, 40 volumes were randomly selected as inputs. All the input volumes were of (110, 251, 187) spatial and of (1, 1, 2) mm voxel resolution. The results of these experiments can be seen on Figure 2. Figure 2 (a) displays the central slices of each of the input volumes before and Figure 2 (b) after the alignment. We can see that after the alignment process the input volumes are nicely aligned.
 |
 |
Medical
We also ran experiments on a real population of MRI acquisitions. The set consisted of 22 baby brain scans of (256,256,124) spatial and (.9375,.9375,1.5) voxel dimension. The results can be seen on Figure 3 and 4. Figure 3 (a) displays the central slice of each of the input volumes before and Figure 3 (b) after the alignment. Three orthogonal views of the mean volumes computed from these datasets is displayed on Figure 4. We can clearly see that after the populations alignment, the data volumes properly line up and the mean volumes have clean and sharp boundaries.
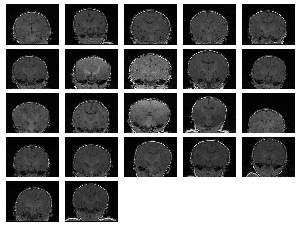 |
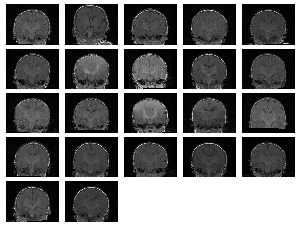 |
 |
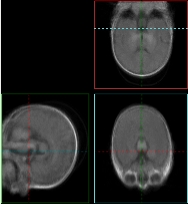 |
Conclusion, Future Work
We introduced a new population registration framework. Without any pre-processing step, we used a congealing-type alignment method to put a large collection of data volumes into correspondence. The algorithm builds on an information theoretic objective function and currently uses affine transformations. The optimization is implemented in a stochastic gradient-based optimization framework that enables a substantial increase in speed.
The promising results of our initial experiments prompt us to further explore the congealing framework. Given that an affine transformation model already produces a close alignment we are now implementing various non-rigid warps that could further refine the image agreement in-between the inputs.
References
[1] E. Miller, N. Matsakis, and P. Viola. Learning from one example through shared densities on transforms. In IEEE Conference on Computer Vision and Pattern Recognition, Vol 1, pp 464-471, 2000.
The Stata Center, Building 32 - 32 Vassar Street - Cambridge, MA 02139 - USA tel:+1-617-253-0073 - publications@csail.mit.edu (Note: On July 1, 2003, the AI Lab and LCS merged to form CSAIL.) |
