Classifying Inter-concept Relations In Medical Discharge Summaries Using Linguistic Information
Tawanda C. Sibanda, Ozlem Uzuner & Peter Szolovits
Motivation
Imagine having the ability to automatically search through medical discharge summaries to find diagnoses and subsequent treatment for patients suffering from shortness of breath and accelerated heart rate and who have benefitted from a particular treatment. Keyword-based techniques can identify documents that mention these concepts but do not differentiate between documents that contain different relations between these concepts, e.g., they can identify patients who have suffered from these illnesses and who have been subjected to the treatment, but cannot separate the patients who benefitted from the treatment from those who have not. We propose a scheme that will allow computers to take a medical discharge summary written in narrative form, label the key concepts in each sentence, and identify the semantic relations between these concepts. Ultimately our scheme can be used to index discharge summaries by their semantic information at the sentence level, facilitating semantic search of these documents.
Related Work
This work is based on previous work by Rosario and Hearst [1]
who describe a scheme that distinguishes seven relation types that can
occur between the entities "disease" and "treatment" in bioscience text.
Rosario and Hearst begin by parsing the text and constructing a vector
of features for each word in the text. They use the following features:
the word itself, its part of speech from the Brill tagger
[2], the phrase from which the word was extracted, the word's MeSH
ID [3], a tri-valued attribute indicating whether the
word is a disease or a treatment or neither (based on MeSH), and the word's
orthographic characteristics (such as capitalization). The authors then
train several Hidden Markov Models and Neural Networks to classify the
disease-treatment relations, using the aforementioned features as inputs.
Using dynamic hidden Markov models, the authors achieve an F-measure of
0.71, where F-measure is calculated using ![]() and PRE is the precision while REC is the recall. Their results
and data show that the most important features for relation classification
are the word itself and its MeSH based attributes.
and PRE is the precision while REC is the recall. Their results
and data show that the most important features for relation classification
are the word itself and its MeSH based attributes.
Approach
Our approach differs from Rosario and Hearst's in three significant ways. Firstly, we concentrate on identifying relations in medical discharge summaries (prior work focused on bioscience text). Discharge summaries present different sets of challenges, as they often consist of ungrammatical sentences and fragments. Secondly, we do not limit ourselves to identifying relations between a single pair of entities. Instead, we have compiled a list of 6 concepts (disease, treatment, symptoms, substances, consult and tests), and 44 relations between pairs of these concepts (e.g., disease causes symptom, treatment cures disease, test confirms disease diagnosis, etc.). These relations capture the majority of the recurring and important semantic information in medical discharge summaries at the sentence level. Finally, we endeavor to determine the linguistic characteristics of the text that are most important in classifying semantic relations. Human beings utilize various information sources, such as words in the sentence and their semantic meaning, the structure of the sentence (its syntax), and the context surrounding the sentence, in inferring semantic meaning. Using Bayesian models, we attempt to find the combination of features that best allows the computer to discern the meaning of a sentence.
The Process
The first step is annotation of the data. For example, consider the following sentence:
"At the time of admission the patient was given Calcium Gluconae and Kayexalate for hyperkalemia."
After annotation this becomes:
"<TAD>At the time of admission the patient was given<med>Calcium Gluconate</med> and <med>Kayexalate</med> for <dis>hyperkalemia</dis></TAD>"
The tags <med> and </med> indicate that the enclosed words represent types of treatment or medication; the tags <dis> and </dis> indicate that "hyperkalemia" is a type of disease; the tags <TAD> and </TAD> represent the relation between the concepts of disease and treatment in the sentence. TAD stands for the relation Treatment Administered for Disease.
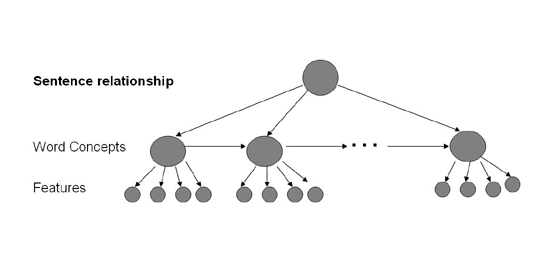
Once all the data has been annotated by a human annotator, a third party tagger (Brill Tagger) is used to identify the parts of speech of the individual words. The tagged output is then preprocessed and fed to a third party parser (Collins' parser [4]) to create a flattened parse tree. We assume that the data can be modeled using a Hidden Markov Model as shown in Figure 1. The top node is the relation evident in the sentence. The second level of nodes represents the roles or concepts in the sentence (such as disease). There is a single concept node for each word in the sentence. Finally the bottom layer of nodes represents the observed features.
Initially, we reproduce Rosario and Hearst's results using five observed features for each role - the word itself, its MeSH ID, the phrase from which the word was extracted, a tri-valued attribute indicating whether the word is a disease or a treatment, and the word's orthographic features. Using these features, we feed the tagged data, the annotated data, and the parse tree to a Perl program that computes the probabilities of the model by simple counting. We use absolute discounting to handle zero counts. To classify a sentence given the model, we extract five features for every word in the sentence and set the evidence variables of the model (the bottom layer) to be equal to these observed values. We then use the junction tree inference algorithm to deduce the most likely relation in the sentence.
Ultimately, our goal is to semantically interpret discharge summaries using linguistic features. In addition to Rosario and Hearst's proposed features, we plan to use syntactic information which we believe will be important. Hence we plan to use features such as the word, the syntactic structure that it occurs in, its semantic type as suggested using ontologies other than MeSH, the syntactic structure of the entire sentence, and the relation of the preceding sentence.
Research Support
This project is funded by "National Multi-Protocol Ensemble for Self-Scaling Systems for Health" (NMESH) contract # N01-LM-3-3515.
References
[1] Barbara Rosario, Marti Hearst, “Classifying Semantic Relations in Bioscience Texts”, Proceedings of the Annual Meeting of Association of Computational Linguistics (ACL ’04), 2004.
[2] Eric Brill, “Transformation-based error-driven learning and natural language processing: A case study of part-of-speech tagging”, Computational Linguistics, 21(4):543-565. 1995.
[3] “Unified Medical Language Systems”, http://www.nlm.nih.gov/research/umls/.
[4] Michael Collins. "A new statistical parser based on bigram lexical dependencies", Proceedings of Annual Meeting of Association of Computational Linguistics (ACL ’96), 1996.
The Stata Center, Building 32 - 32 Vassar Street - Cambridge, MA 02139 - USA tel:+1-617-253-0073 - publications@csail.mit.edu (Note: On July 1, 2003, the AI Lab and LCS merged to form CSAIL.) |