Engineering In Vivo Transcription-Based Digital Logic
Reshma P. Shetty & Thomas F. Knight, Jr.
Synthetic Biology
Synthetic Biology is an emerging engineering discipline whose goal is the rational design, fabrication and analysis of systems built from biological parts. Just as electrical engineering has its foundations in physics, synthetic biology seeks to make use of biological science to develop new technologies. See the website for more details.
Information processing via transcription-based logic
One class of synthetic biological systems are those whose purpose is to process information. My work seeks to build transcription-based devices in vivo capable of carrying out combinational digital logic. Devices based on proteins which regulate transcription from DNA are attractive candidates for engineering because protein-DNA interactions are relatively well understood.
The figure below shows a simple logic device, the inverter, in typical digital logic (left) and in transcription-based digital logic (right). In contrast to the typical digital logic device which represents signals in voltages (V), the transcriptional inverter carries its signals in transcription rates. It has input signal πi and output signal πo. In the transcription-based device, a gene encodes a protein (block arrow) capable of binding to a promoter (line arrow) and regulating transcription from that promoter. Thus, a high input transcription signal to the gene causes lots of protein to be made. That protein can then bind to the promoter and repress transcription thereby causing inversion of the input signal πi.
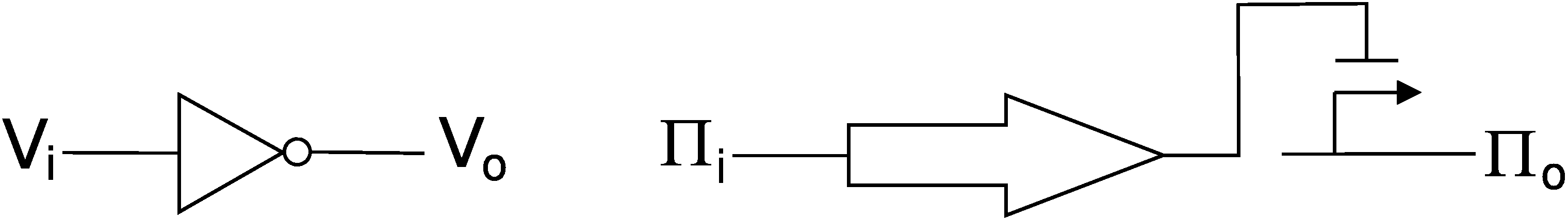
There are currently several key obstacles to the goal of transcription-based digital logic. First, the current implementation of transcription-based logic is limited to only a few devices. Second, existing devices are poorly characterized making it difficult to use them in larger systems. Third, existing devices cannot be cascaded easily since their signals are not well matched.
Device parameterization
Standardized protocols for characterization of transcriptional devices do not exist. To engineer composable devices, it is necessary to measure and characterize individual device behavior. We are in the process of developing methods for measuring the static device behavior (or transfer curve) of these transcription-based devices. These methods are being developed using existing devices but should be readily applicable to novel devices as well.
Develop zinc finger and leucine zipper technology for modular, synthetic transcription factors
Most existing devices are derived from bacterial repressors which are limited in number and do not necessarily function as intended in transcriptional devices. We will use well-studied zinc finger and leucine zipper protein domains to build modular transcription factors. The modular design of these transcription factors will facilitate the development of tunable parts and add scalability to transcription-based logic. Currently, we have developed simple physicochemical models of these devices to inform device design. Information from the model is being used to guide design of synthetic transcription factors.
Funding
Funding for this work has generously been provided by the National Science Foundation Graduate Research Fellowship and the Whitaker Foundation Graduate Fellowship.
References:
[1] T. S. Gardner, C. R. Cantor and J. J. Collins. Construction of a genetic toggle switch in Escherichia coli. In Nature, 403(6767):339-42, 2000.
[2] J. R. S. Newman and A. E. Keating. Comprehensive identification of human bZIP interactions with coiled-coil arrays. In Science, 300(5628):2097-101, 2003.
[3] R. Weiss. Cellular Computation and Communications using Engineered Genetic Regulatory Networks. PhD thesis, Massachusetts Institute of Technology, 2001.
[4] S. A. Wolfe, E. I. Ramm and C. O. Pabo. Combining structure-based design with phage display to create new Cys2His2 zinc finger dimers. In Structure, 8(7):739-50, 2000.
[5] S. A. Wolfe, R. A. Grant and C. O. Pabo. Structure of a designed dimeric zinc finger protein bound to DNA. In Biochemistry, 42(46):13401-9, 2003.
The Stata Center, Building 32 - 32 Vassar Street - Cambridge, MA 02139 - USA tel:+1-617-253-0073 - publications@csail.mit.edu (Note: On July 1, 2003, the AI Lab and LCS merged to form CSAIL.) |
