The Molecule Self-Reconfiguring Robot
Keith Kotay & Daniela Rus
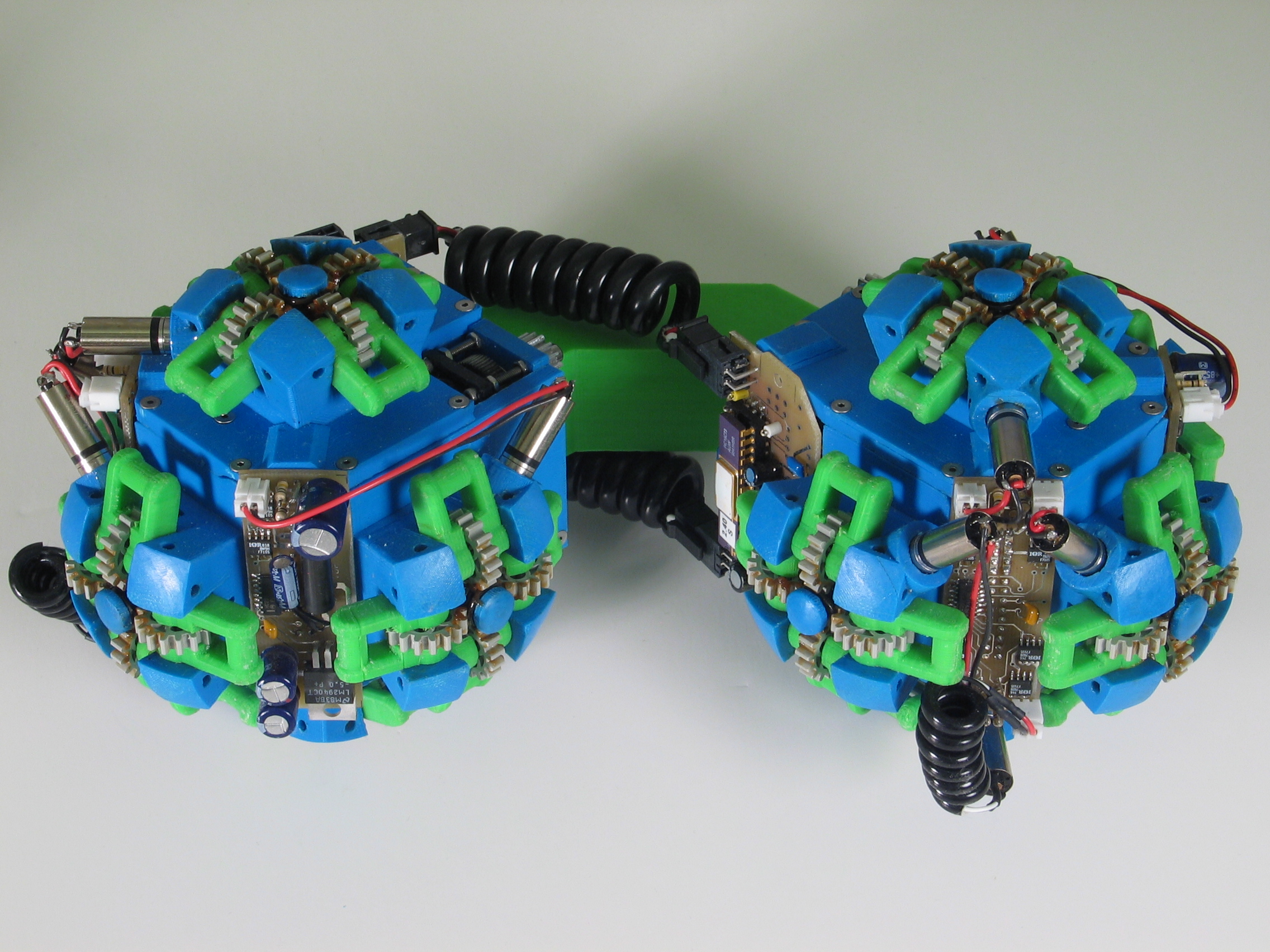
Fig. 1: The two types of Molecule modules. The male Molecule (left) has the active gripper mechanism, and the female Molecule (right) has the passive fixture which is grabbed by the gripper arms.
Background
Self-reconfiguring robots are modular robots that are physically connected and can achieve structural geometric changes autonomously. Modular robots are well-suited for locomotion in unknown environments. For example, self-reconfiguring robots can climb stairs even in the absence of models of the height, width, and length of the stairway. The robot will be given the command to move forward. The robot will proceed with a translation motion until the front sensors mounted on the forward modules detect an obstacle (e.g., the first step). At this point the robot will change the locomotion modality from translation to stacking. When the top modules detect free space again (that is, after the first step has been cleared), the robot will change locomotion modality again to unstacking and then translation. These capabilities lead to on-line algorithms for navigation that take advantage of self-reconfiguring capabilities to create a "water-flow"-like locomotion.
In [1] we describe a generic algorithm for implementing the "water-flow" locomotion gait using the self-reconfiguration property. In [2] we instantiated the algorithm to the Molecule robot [3-8]. We described a distributed statically stable algorithm for the "water-flow" locomotion gait using the specific degrees of freedom of Molecule modules. In this gait a group of modules tumble on top of each other to implement forward progress. We analyzed the efficiency of this gait in terms of the number of actuations, and uses this result as inspiration to develop a rolling gait which is dynamically but not statically stable. Using self-reconfiguration, a group of modules can actively change their center of mass to generate forward motion. We analyzed the efficiency of this gait and showed that the dynamically stable algorithm is more efficient. We also demonstrated the locomotion tumbling gait on a four-module Molecule robot and discussed our experimental data.
Molecule Module Description
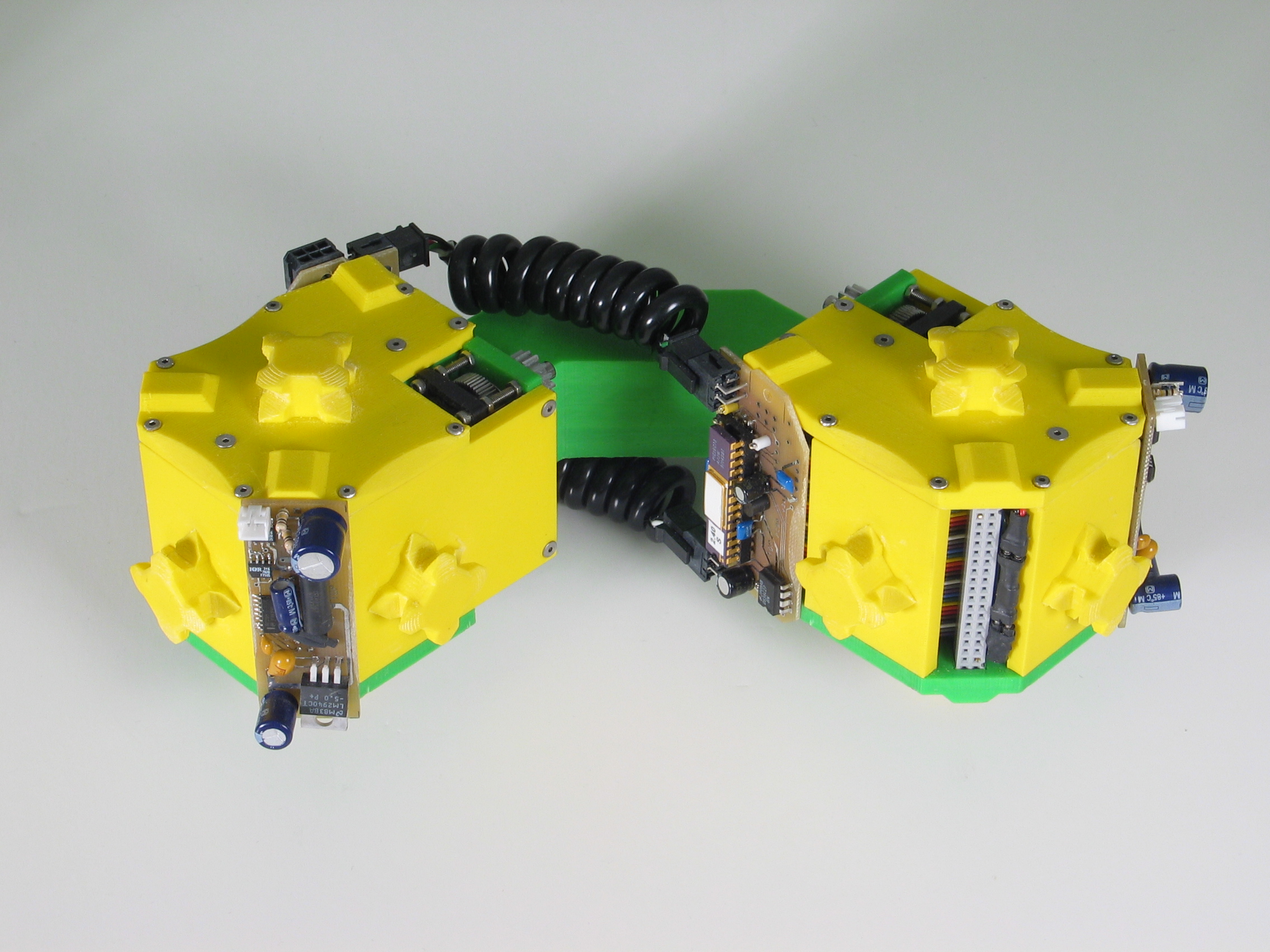
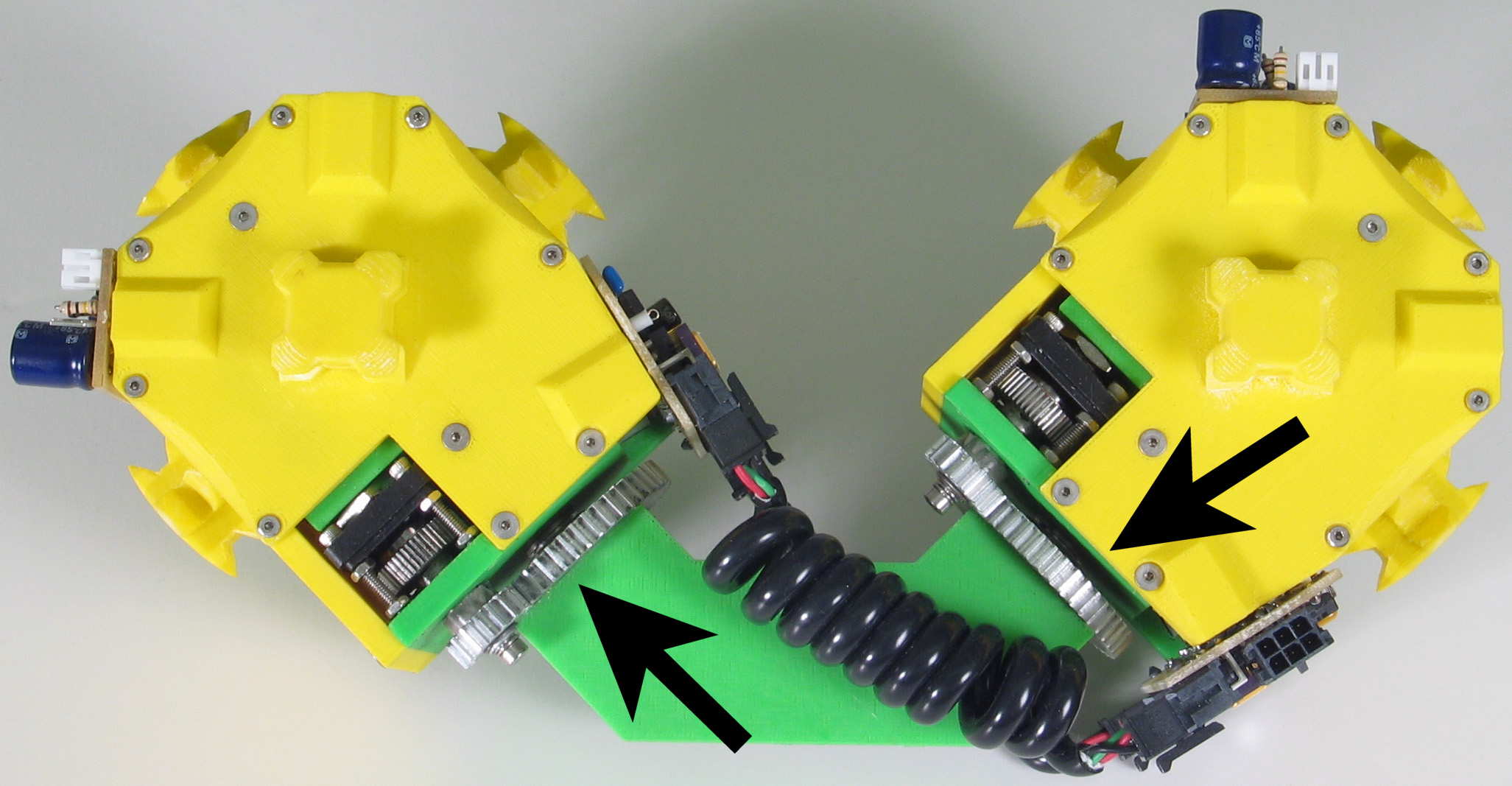
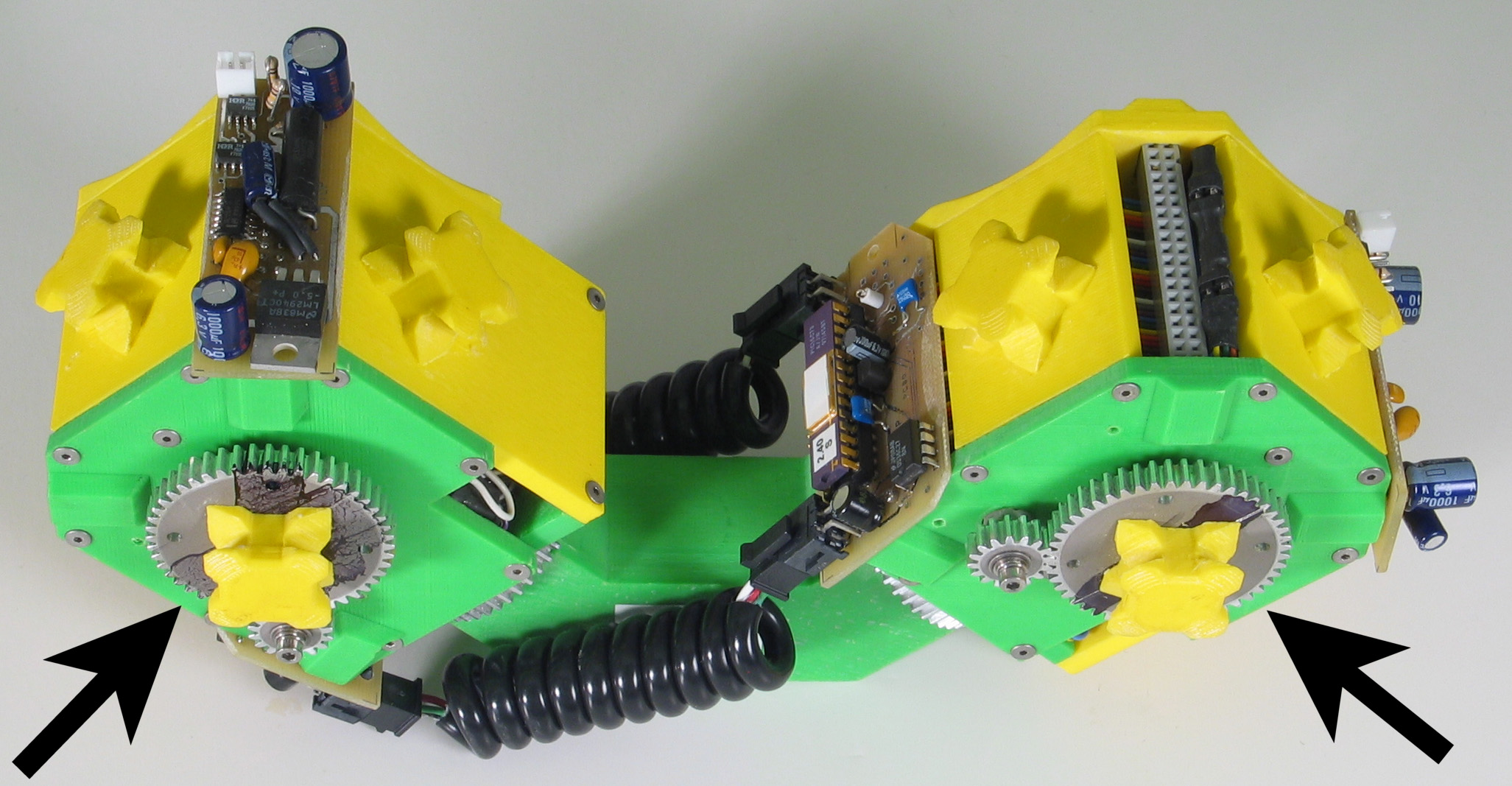
The left image indicates the bond degrees-of-freedom, and the right image indicates the connector degrees-of-freedom.
The Molecule is a robotic module capable of aggregating with other identical modules to form dynamic three-dimensional structures. Molecules can selectively form rigid connections to other Molecules and, by using these connections and their rotating degrees of freedom, modules can move to different locations on Molecule structures. A Molecule module is composed of two cubical atoms linked by a rigid 90-degree link called the bond (see Fig. 1). Each atom has five inter-Molecule connection points and two degrees of freedom (see Fig. 2). One degree of freedom allows the atom to rotate 180 degrees relative to its bond connection, and the other degree of freedom allows the atom (and therefore the entire Molecule) to rotate 180 degrees relative to one of the inter-Molecule connectors. The bond and connector degrees of freedom permit independent movement on a substrate of identical Molecules, including straight-line traversal and 90 degree concave and convex transitions to adjacent surfaces. A Molecule moves by attaching an atom to some other Molecule and actuating one or more of its four degrees of freedom. Attachments are made using the gripper connector mechanism, which consists of an active four-armed gripper and a passive "grippee" which provides an anchoring surface for connection.
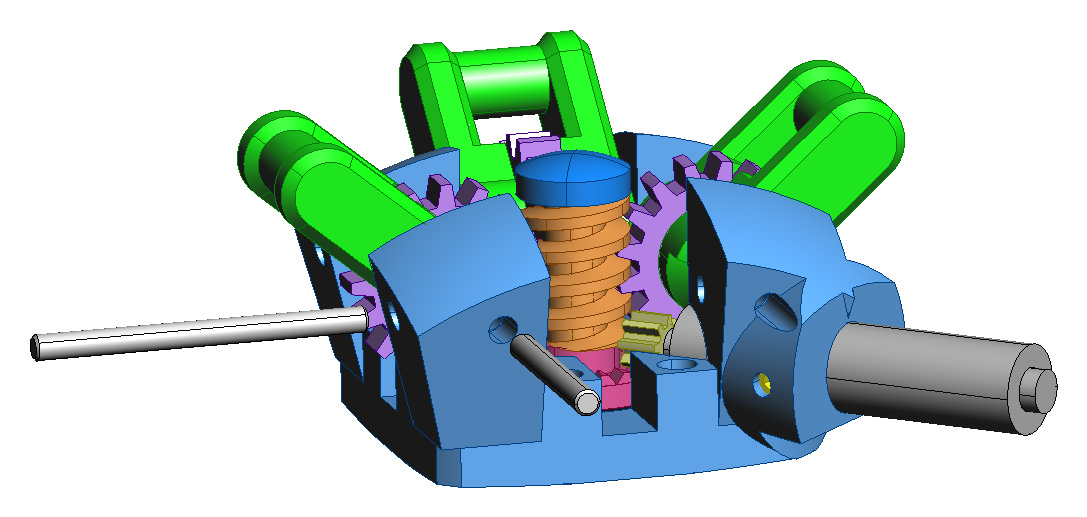
Molecule Design
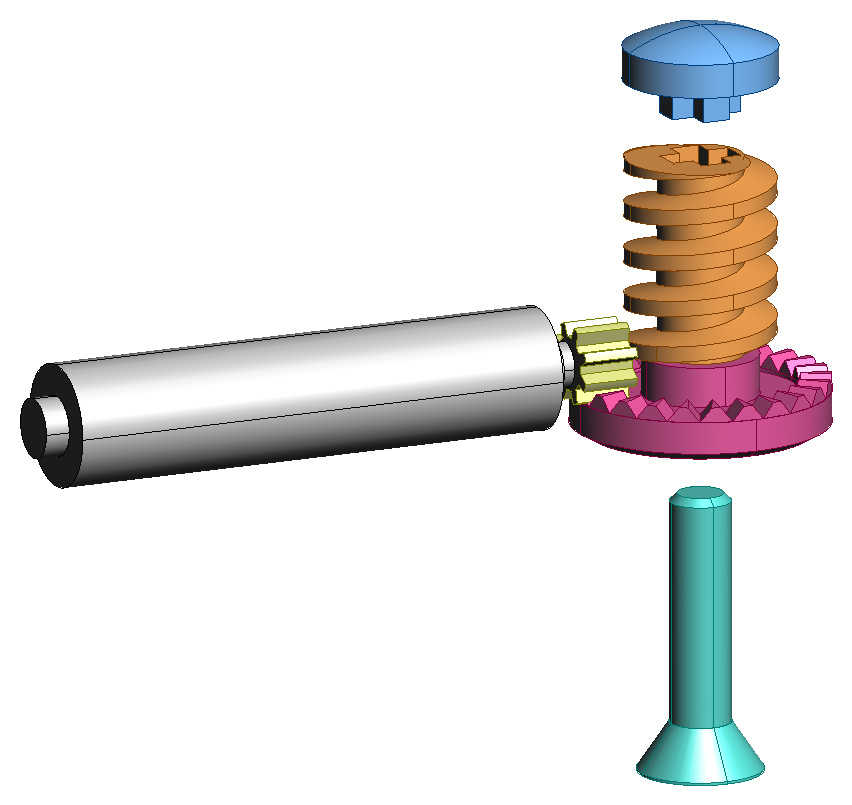
Fig. 3: CAD drawings of the gripper
assembly. In the left image the north arm has been removed to show the
internal configuration.
The north and east arm shafts are shown in exploded view. The right image
shows the drive train in detail,
with the shaft (a flat head machine screw) and worm gear cap shown in
exploded view.
Our design goal for the Molecule was two-fold: (1) creating a module that could independently relocate on a static module substrate, and (2) reducing the required actuation hardware to the absolute minimum to minimize module size and weight. Given a cubic lattice and a face-to-face connection scheme, it is easy to see that three movement modalities are sufficient to traverse any structure: translation, concave transition, and convex transition. Concave transition is the simplest of these modalities---a single cube in a lattice automatically spans the concave transition formed by two neighboring cubes. Translation involves moving from one cube face to another cube face, where both faces are in the same plane. In a system with discrete connection points, a module must span the distance between the two faces. Convex transition is the most demanding modality since the module must span the distance from one face of a cube to a neighboring face on the same cube. This can be achieved by using an ``L''-shaped module occupying three lattice positions. Since an ``L''-shape is also capable of concave transitions and translations it is sufficient for independent lattice traversal.
However, simply composing three cubical units would require large actuation forces due to the combined weight of the modules and the distance between the units at the ends of the ``L''-shape (atoms). The simplest optimization is to replace the center module with a rigid link (bond). The atoms are required for connection during a convex transition, and are also sufficient for translations and concave transitions. Further analysis shows that each of the atoms require only two degrees of freedom: (1) rotation about the bond, and (2) rotation of one of the connection points whose face is adjacent to the face which connects to the rigid link. Both degrees of freedom require a minimum rotation of 180 degrees. The result is a module with four rotating degrees of freedom and ten connectors.
The Molecule gripper connector is a binary connector with 90-degree symmetry, a large amount of compliance, and the ability to retract within the bounding sphere of the atom which permits in-place atom rotations. Fig. 3 (left) shows the active male part of the gripper system which contains the motor, gears, and the four arms which rotate into place to grip the passive female part. The arm subassemblies surround a central worm gear which turns to actuate the arm gears. The worm gear is on the same shaft as a crown gear which is turned by the gearmotor, as shown in Fig. 3 (right). The gearmotor has a diameter of 8 mm and an integral 256:1 geartrain.
Molecule Robot Locomotion
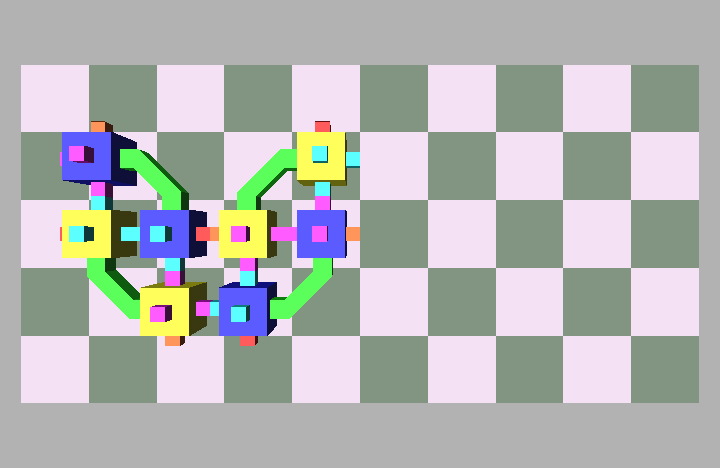
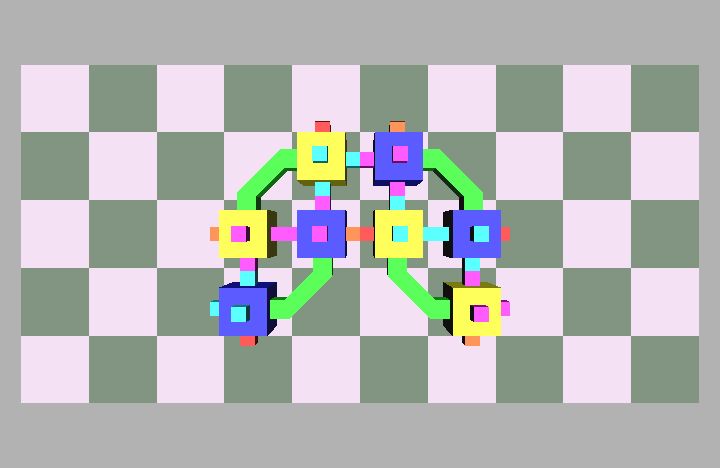
Fig. 4: Five snapshots taken from a Molecule pair translation simulation utilizing two Molecule pairs. The motion is in the +x direction.
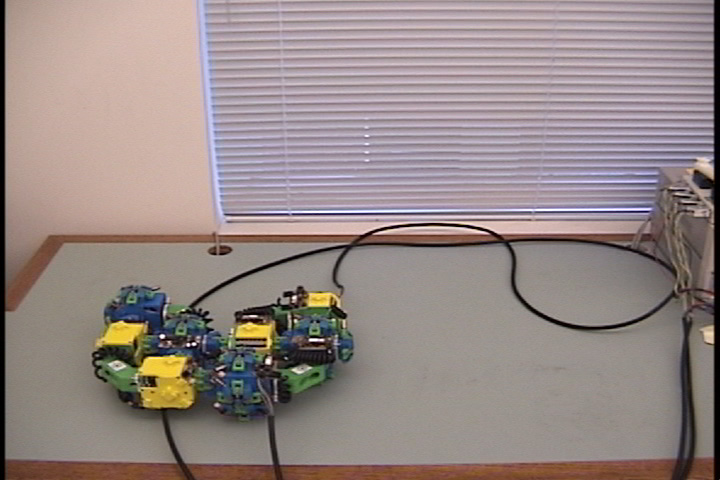
Molecule structures move by dynamically changing the shape of the structure. Locomotion entails the relocation of modules in the direction of motion while maintaining a stable structure. For example, if modules at the rear of the structure move to the front then the structure achieves a net displacement. Fig. 4 shows a simulation of two Molecule pairs moving from the right to the left side of the group, effectively translating the structure. This process can be repeated by continually moving the rearmost pair to the front, allowing for arbitrary translation distances. This algorithm works for an arbitrary size robot as described in [8]. Fig. 5 shows the Molecule pair translation experiment on actual hardware.
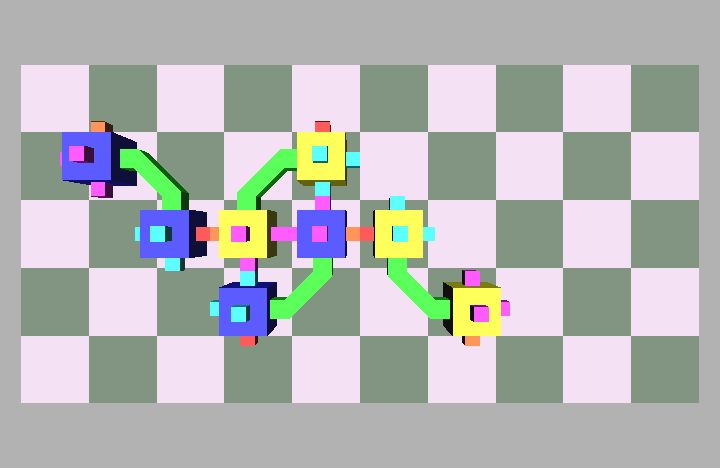
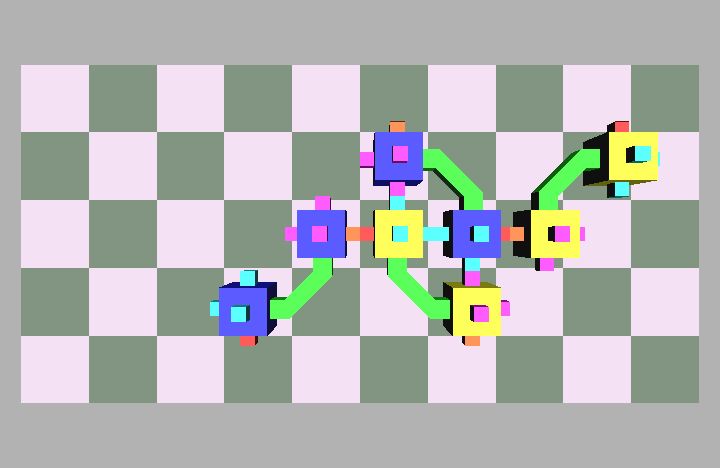
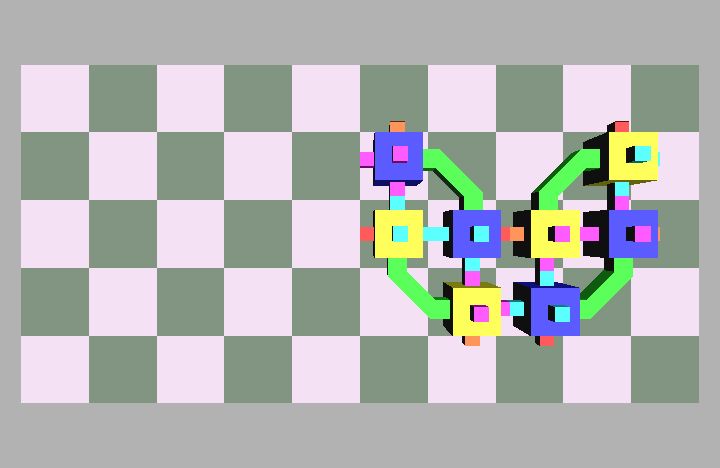
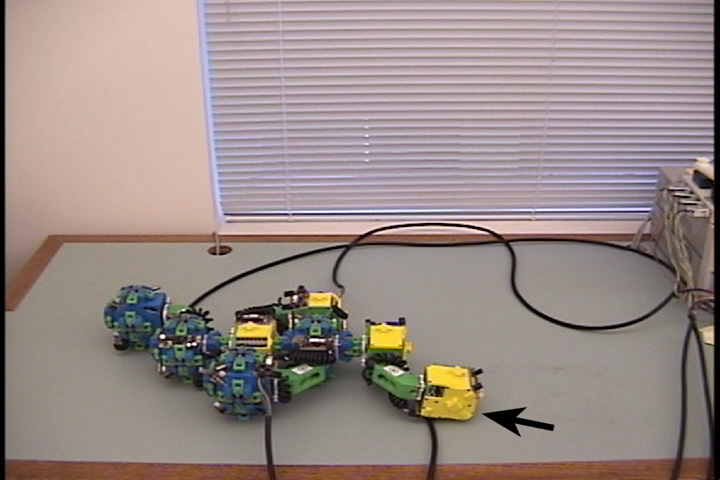
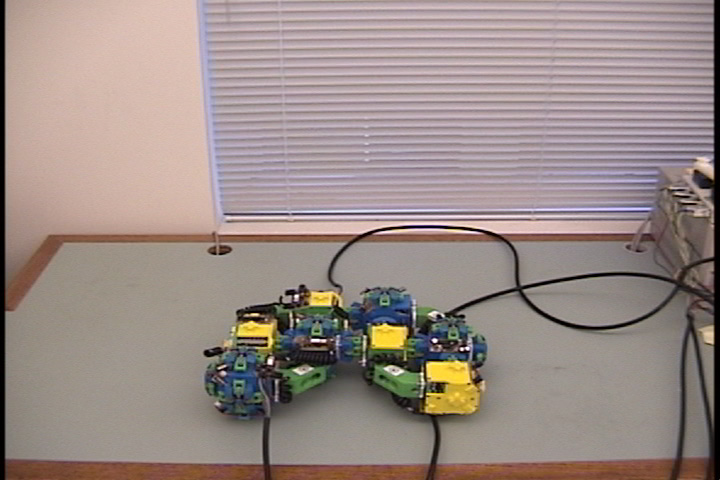
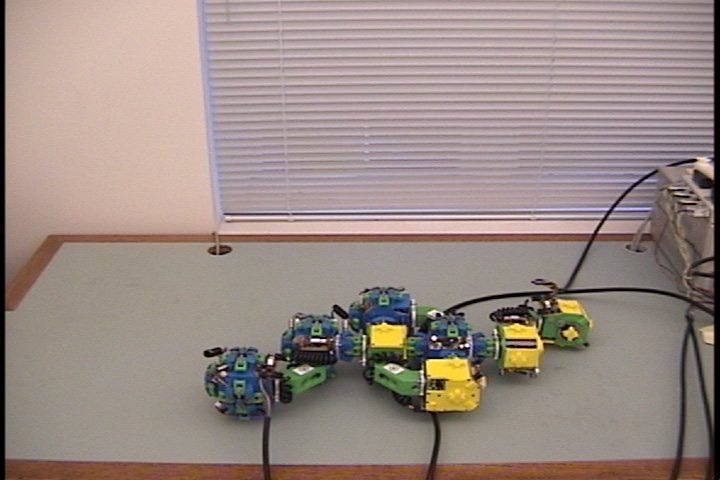
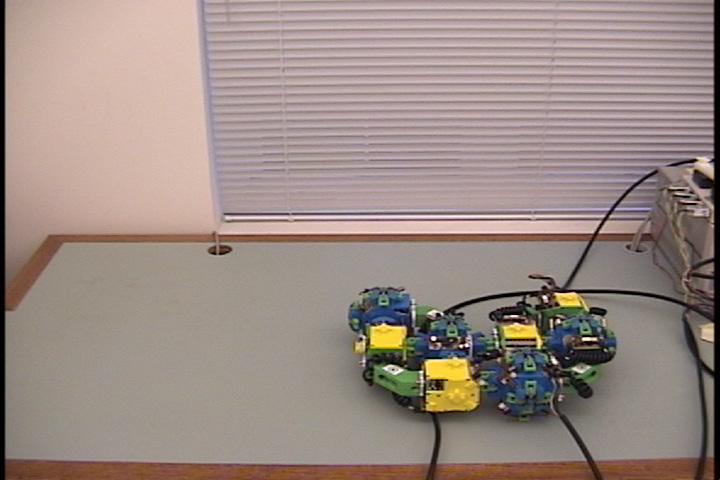
Fig. 5: Five snapshots from a Molecule
pair translation experiment utilizing two Molecule pairs. The motion is
in the +x direction.
The arrow in the second image shows the female atom lowered into contact
with the table to increase the stability of the structure.
Research Support
Support for this work was provided through NSF awards IRI-9714332, EIA-9901589, IIS-9818299, IIS-9912193 and EIA-0202789 and ONR award N00014-01-1-0675. We are grateful for this support.
References:
[1] Zack Butler, Keith Kotay, Daniela Rus, and Kohji Tomita. Generic Decentralized Locomotion Control for Lattice-Based Self-Reconfigurable Robots, Intl. Journal of Robotics Research, vol. 23, no. 9, 2004.
[2] Keith Kotay and Daniela Rus. Efficient Locomotion for a Self-Reconfiguring Robot, Proc. of the IEEE Intl. Conf. on Robotics and Automation, 2005.
[3] Keith Kotay, Daniela Rus, Marsette Vona, and Craig McGray. The self-reconfiguring robotic Molecule: design and control algorithms, Proc. of the Workshop on the Algorithmic Foundations of Robotics, 1998.
[4] Keith Kotay, Daniela Rus, Marsette Vona, and Craig McGray. The Self-reconfiguring Robotic Molecule, Proceedings of IEEE Intl. Conf. on Robotics and Automation, 1998.
[5] Keith Kotay and Daniela Rus. Motion Synthesis for the Self-reconfiguring Molecule, Proc. of the Intl. Conf. on Intelligent Robots and Systems, 1998.
[6] Keith Kotay and Daniela Rus. Locomotion versatility through self-reconfiguration, Robotics and Autonomous Systems, vol. 26, pp. 217--232, 1999.
[7] Keith Kotay and Daniela Rus. Algorithms for Self-reconfiguring Molecule Motion Planning, Proc. of the Intl. Conf. on Intelligent Robots and Systems, 2000.
[8] Keith Kotay. Self-Reconfiguring Robots: Designs, Algorithms, and Applications, Ph.D. Thesis, Dartmouth College, Computer Science Department, 2003.
The Stata Center, Building 32 - 32 Vassar Street - Cambridge, MA 02139 - USA tel:+1-617-253-0073 - publications@csail.mit.edu (Note: On July 1, 2003, the AI Lab and LCS merged to form CSAIL.) |