From Spatial Regularization to Anatomical Priors in fMRI Analysis
Wanmei Ou & Polina Golland
Background and Problem
Functional Magnetic Resonance Imaging (fMRI) has played a critical role in psychological research and clinical practice since its development in the early 1990s. fMRI techniques were developed based on Magnetic Resonance Imaging (MRI), which generates pictures of soft tissue such as the brain, in a static state for anatomy identification. It has been discovered that neural activation causes incremental blood flow. fMRI analysis utilizes this fact to generate a series of pictures over time indicating neural activation, thus enabling studies of the brain in a dynamic state. It provides us a discrete time signal for each location of the brain with a sampling rate on the order of one second. With carefully designed psychological experiments, we can understand the neural basis of our behavior from simple to cognitively complex functions. Simple tasks include movement of fingers and basic sensory perceptions in response to stimuli based on presentation of light, tones, and syllables. More complicated tasks include face recognition, memory, and fear. The psychological experiments aim to functionally associate one or more brain regions with the task a subject performs.
Modeling brain activation based on fMRI has been an active research area since its development. A subject undergoes fMRI scans while alternating between performing tasks and resting. Neuroscientists have developed various algorithms to determine which locations of the image are active. From this information an activation map is produced. The activation map indicates points in the image where the brain has become active due to a stimulus. These algorithms, operating on each location of the brain separately, measure the difference in the signal obtained between the task and rest periods. Due to a lack of knowledge of brain activities, researchers usually apply arbitrary thresholds to the activation map that each algorithm generates. Locations with statistics above the threshold are considered active; otherwise, not active. The major disadvantage of fMRI is a low signal-to-noise ratio which makes detection difficult. Using arbitrary thresholds results in a number of scatter activation islands in the activation map; most of which are ignored in further analysis and interpretation. This approach reduces the accuracy of the results, and therefore is a crucial area which needs to be improved.
Our Solution
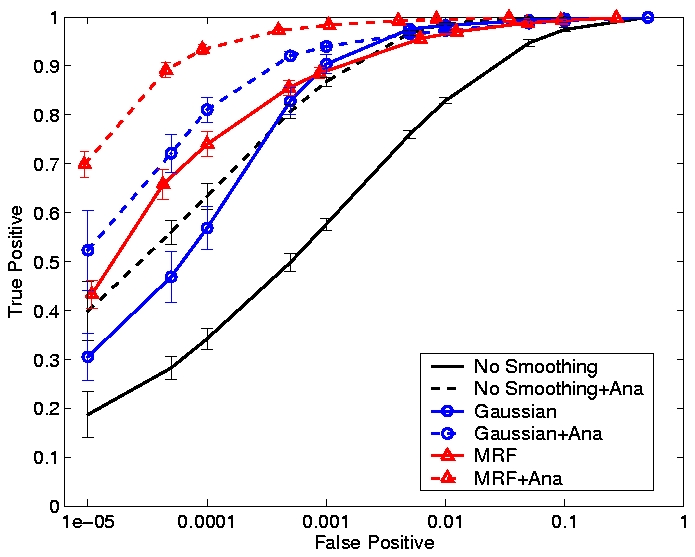
According to fMRI image properties, neighboring locations tend to be at the same activation state. We can take advantage of this characteristic to reduce scatter activation islands. A common approach to reducing such false detections employs a Gaussian filter to smooth the fMRI signal prior to applying a detector. We refer this process as the Gaussian-smoothing-based detector. Unfortunately, Gaussian smoothing, though intended to combat low SNR, leads to a loss of detail in the resulting binary activation maps. Conventional image processing research has proved that Markov random field (MRF) can overcome this issue. We adopted MRF as our spatial model. It is difficult to verify the model accuracy since the ground truth of brain activation is unknown. We therefore used simulated activations with a realistic noise model, extracted from true fMRI images, to study the behavior of MRF-based detectors at different noise levels. The true positive / false positive rate, also unknown as receiver operating characteristic (ROC) analysis, is the measurement of the detectors performance. Compared with Gaussian smoothing in Fig.1, the MRF-based detector achieves a ten percentile higher true positive rate when false positive rate equals 0.05%. Detailed results can be found in [2].
Inspired by atlas-based segmentation [3], we further improve the performance of the MRF-based detector by incorporating spatial guidelines provided by anatomical images such as MRI. We can segment the anatomical images into different tissue types such as gray matter, white matter and bone. It is known that activation most likely occurs in gray matter. Our goal is to integrate this information into my current work to achieve a higher detection rate. Intuitively speaking, we want the model to reflect the fact that activation is much more likely to occur in gray matter than in white matter, and not at all in bone. In addition, the spatial coherency of activation is strong within each tissue and not across tissue boundaries. The extended MRF model encodes both tissue type and activation state. On the other hand, it is straightforward to inject anatomical information into the Gaussian-smoothing model. We are intreseted at improvement in detection accuracy provided by the anatomical information. Details of this work can be found in [2].
Validation
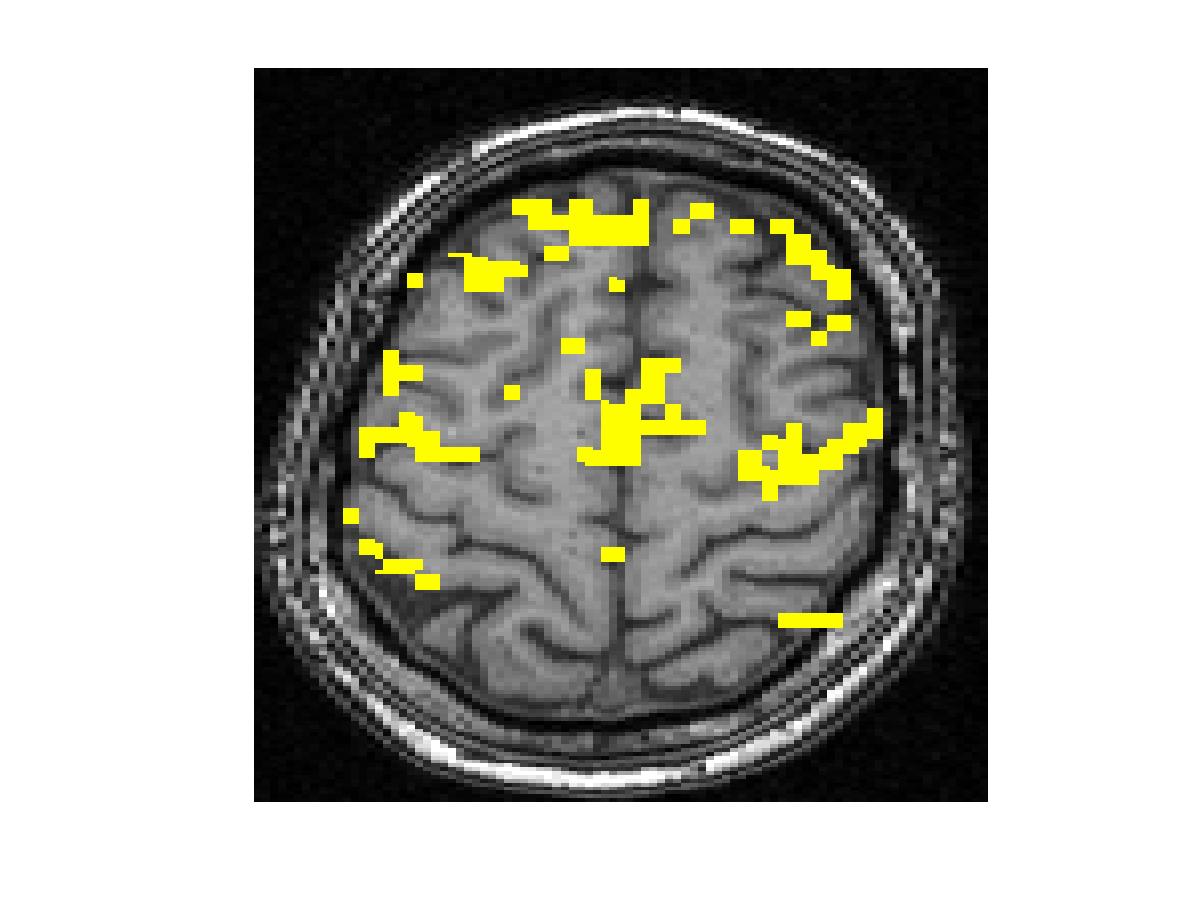
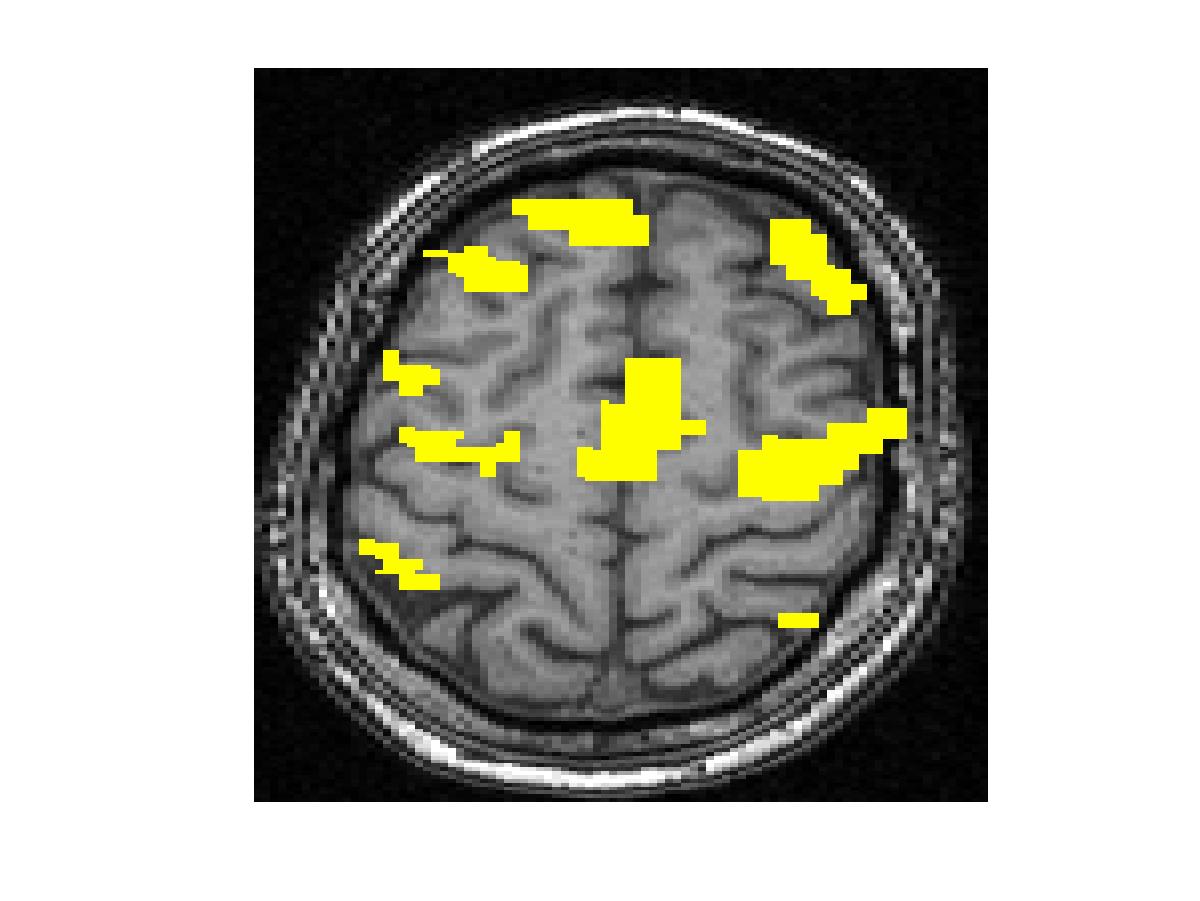
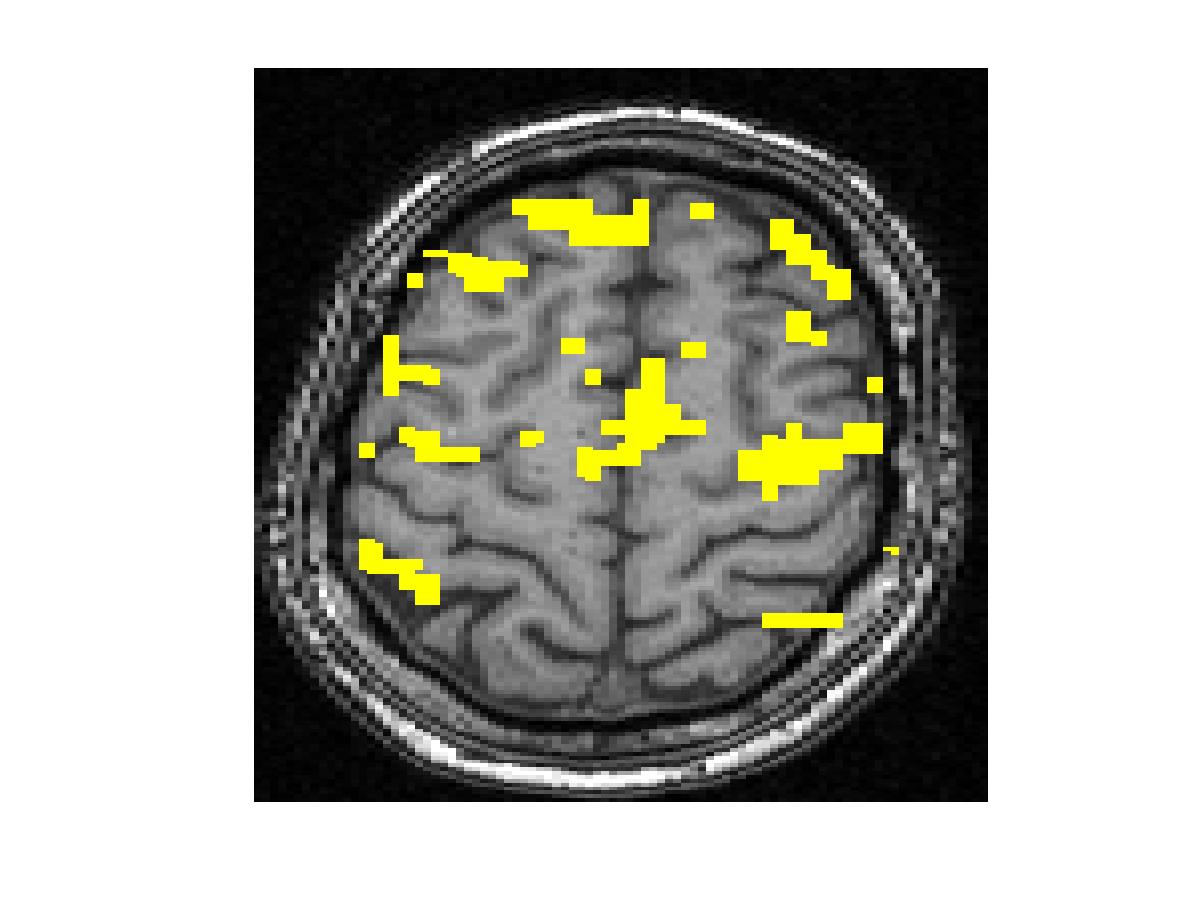
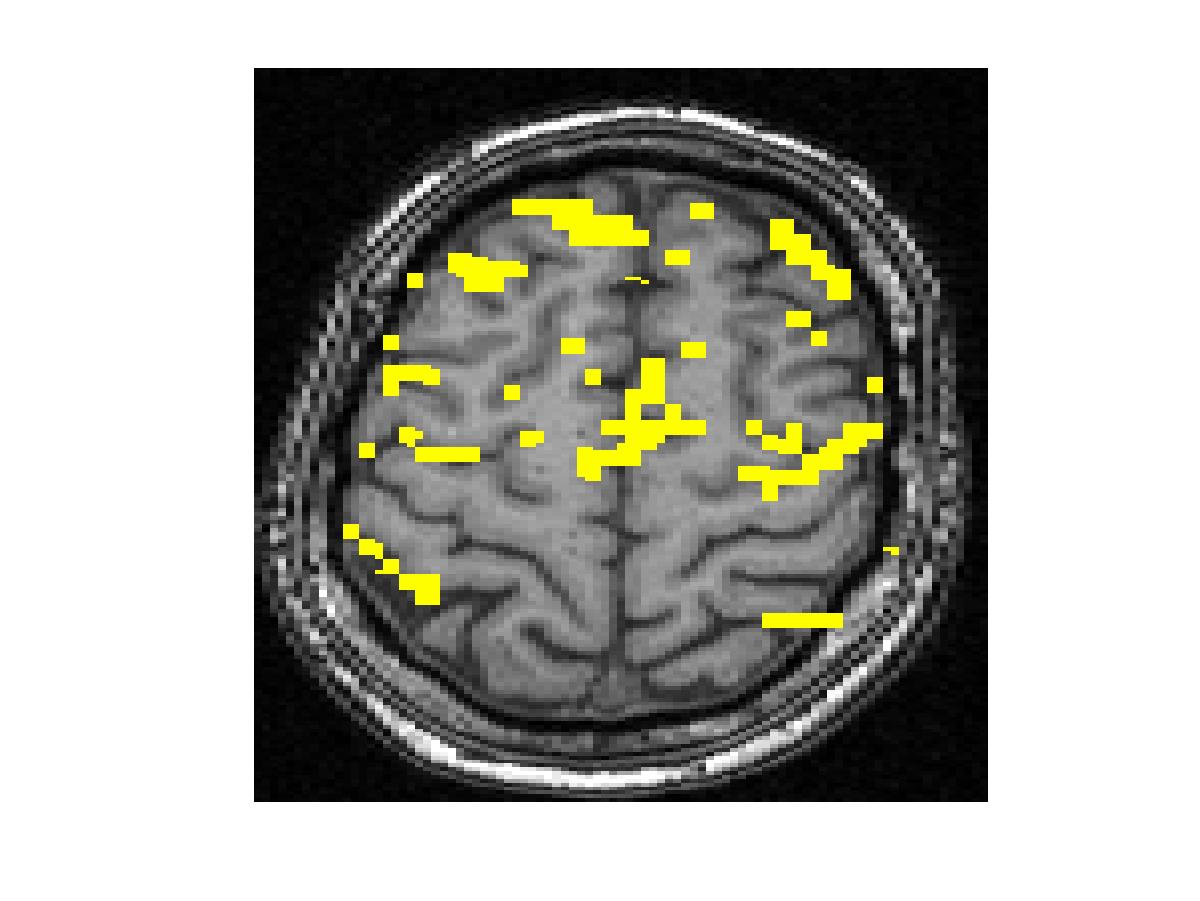
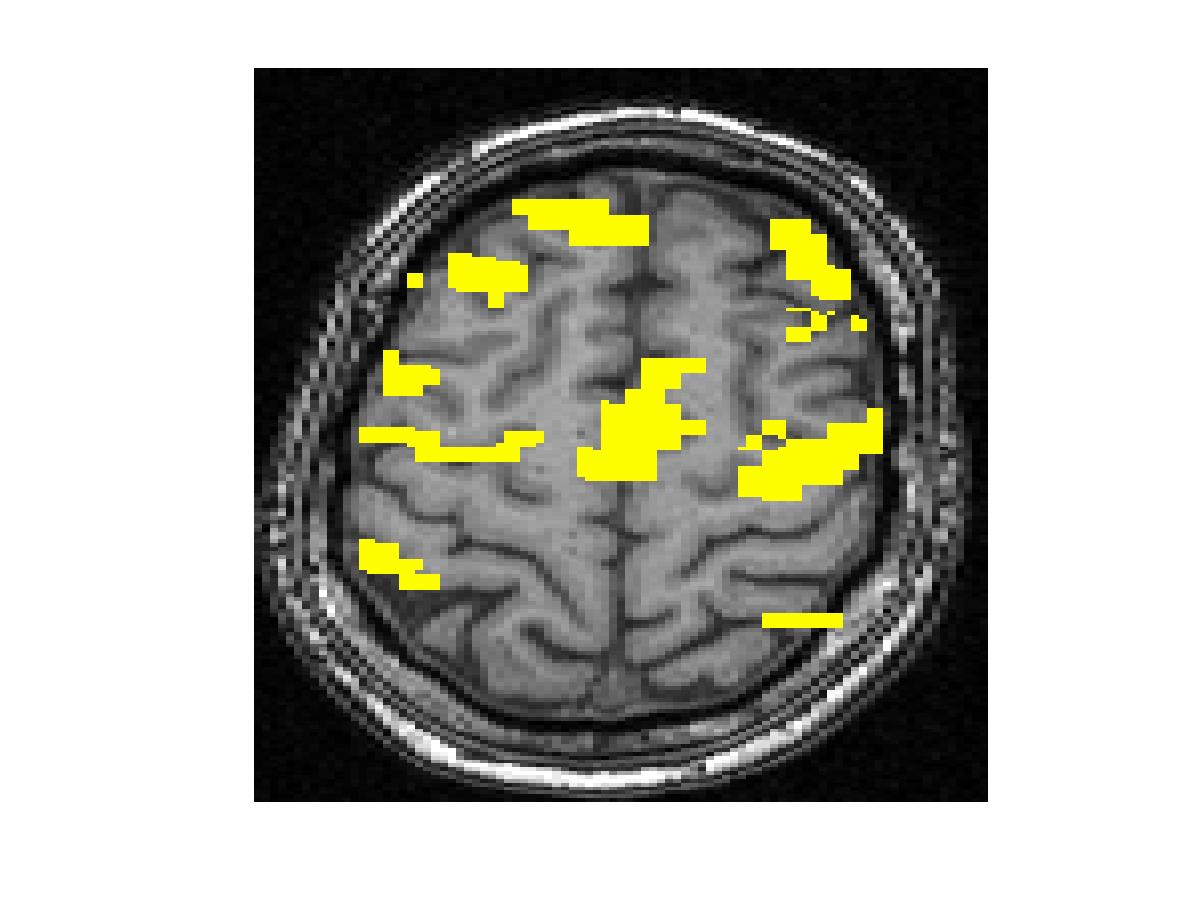
To compare our detectors with other detectors, we performed ROC (Fig. 1) analysis on synthetic data. Injecting anatomical information improves detection accuracy for both the Gaussian-smoothing-based detector and MRF-based detector. We also validated them by studying their ability to recover activation from significantly shorter time course using real fMRI data (Fig.2). (b) shows the result of applying the same detector as that employed in (a) to the first half of the time course. This map is more fragmented due to loss in SNR from reducing the length of the signal. The other four images illustrate the results of applying the same detector with the Gaussian smoothing and the MRF model, as well as their anatomically augmented versions. Although Gaussian smoothing removes most of the single voxel activation islands, its activation map (c) is an overestimate compared with (a). Anatomical information slightly reduces the overestimate in the Gaussian smoothing. the MRF-based detectors (e and f) yield reconstruction results that are close to the activation map estimated from the full-length signal, but do not look overly smoothed. This highlights the potential benefit of using the Markov model in fMRI detection. Similarly to the Gaussian smoothing, the MRF model benefits from using anatomical information to remove spurious activations.
Figure 1. ROC curves for different smoothing techniques at SNR=-6dB. False positive rate is shown on the log scale.
|
| ||
| 
|  |
| 
| 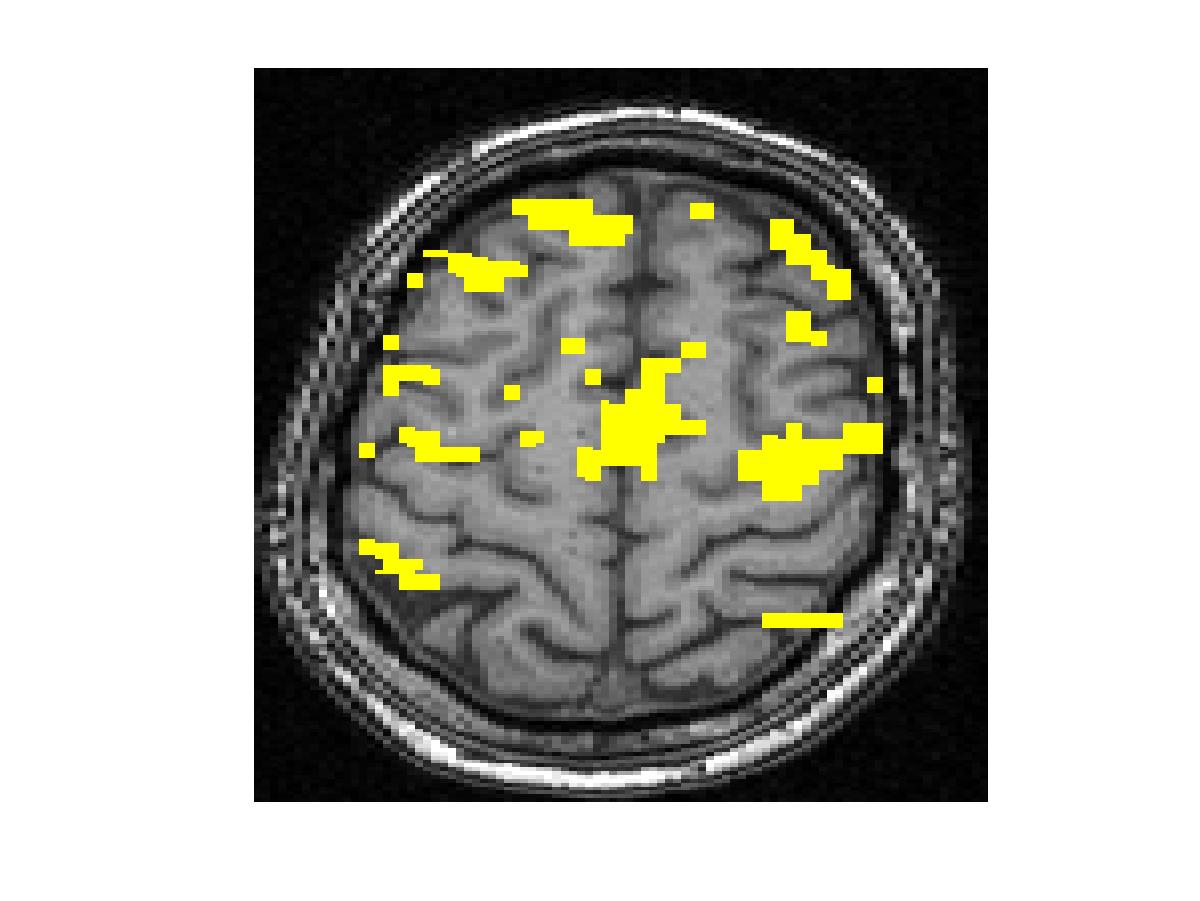 |
Figure 2. Real fMRI study. One slice in the estimated activation map. (a) No spatial smoothing, using the entire time course. (b)-(f) Estimation based on the first half of the time course using different spatial smoothing methods.
Acknowledgements
This work was partially supported by the NIH National Center for Biomedical Computing Program, National Alliance for Medical Imaging Computing (NAMIC), Fund No. 1U54 EB005149, the NSF IIS 9610249 grant, and the NIH R01 NS37922 grant.
References
[1] Jezzard, F., Matthews, P.M. and Smith, S.M. Functional MIR -- An Introduction to Methods. OXFORD, Month 2002.
[2] Ou, W. and Golland, G. From Spatial Regularization to Anatomical Priors in fMRI Analysis. In Proc. IPMI'05, July 2005.
[3] Pohl, K., Incoperating Non-rigid Registration into Expectation Maximization Algorithm to Segment MR Images.In Proc. MICCAI'02, pp. 564-572.
The Stata Center, Building 32 - 32 Vassar Street - Cambridge, MA 02139 - USA tel:+1-617-253-0073 - publications@csail.mit.edu (Note: On July 1, 2003, the AI Lab and LCS merged to form CSAIL.) |