
| Research Abstracts Home | CSAIL Digital Archive | Research Activities | CSAIL Home |
![]()

|
Research
Abstracts - 2007
|
Struct2Net: Integrating Structure Into Protein-Protein Interaction PredictionRohit Singh, Jinbo Xu & Bonnie BergerIntroductionWe consider the problem of predicting if two proteins interact, given their sequence information and, optionally, other genomic and proteomic information. Such computational prediction of protein-protein interactions (PPIs) can supplement experimental methods for elucidating PPIs. When mapping very large interactomes (e.g., human), such PPI predictions--- even if only partially accurate--- would be valuable in prioritizing the set of interactions to experimentally test. Moreover, experimental techniques are quite error-prone; as prediction methods (like the one proposed here) gain accuracy, they can be used to double-check the results of the experiments. MethodsWe present a framework for predicting protein-protein interactions (PPIs) that integrates structure-based information with other functional annotations, e.g. GO, co-expression and co-localization, etc. We use structure-based methods, in conjunction with high-throughput information, to predict interactions. We describe a fully-automated structure-based method for computing the likelihood of an interaction, solely from sequence data. A key idea here is that if a potential interaction is sufficiently favorable energetically, it is likely to be true. As part of our method, we introduce a novel algorithm for computing the most-likely structure of the complex formed by two given proteins and describe the use of logistic regression (see Figure 1) for evaluating if the putative complex corresponds to a true interaction. Furthermore, to the best of our knowledge, this work is the first to describe a framework for predicting PPIs that integrates these structure-based insights with other functional annotation (e.g., co-expression, GO description). We use Random Forests, a recently introduced machine learning technique for the classification. 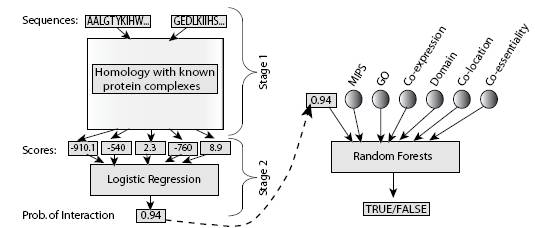 Figure 1: Schematic of Struct2Net ResultsWe tested our method on a set of approximately 1000 yeast genes. The results indicate that the use of structure-based methods can significantly improve our ability to predict PPIs. Interestingly, the predicted interaction network is a scale-free network. Our method also predicted some potential interactions involving yeast homologs of human disease-related proteins. References[1] Rohit Singh*, Jinbo Xu* and Bonnie Berger. Struct2Net: Integrating Structure Into Protein-Protein Interaction Prediction. In The Proceedings of the 11th Pacific Symposium On BioComputing, Hawaii 2006 (*: contributed equally) |
||||
|